Start where you're curious.

May 2019 to May 2025
Pascal Lab
EXPANDING THE LEXA BACTERIAL TWO-HYBRID SYSTEM FOR HUMAN PROTEIN-PROTEIN INTERACTION SCREENING: APPLICATION TO TUMOR SUPPRESSOR PAR-4
My doctoral research focused on expanding the utility of the LexA-based bacterial two-hybrid (B2H) system to study interactions between human proteins, with a specific application to the tumor suppressor Par-4. This work bridges molecular biology and synthetic biology, demonstrating how a prokaryotic system can be adapted for cost-effective, scalable screening of protein–protein interactions relevant to human disease pathways.
Project One, highlighted below, presents the first empirical evidence of a direct interaction between GRP78 and cleaved Par-4 (cl-Par-4), a relationship implicated in apoptosis signaling. This study also marks the first application of the LexA-B2H system for analyzing human protein–protein interactions, demonstrating its potential as a cost-effective, prokaryotic platform for biomedical discovery.
A full copy of the dissertation and public defense is available at the end of this section for readers interested in the complete methodology, data, and broader research implications.

Domain Structure of Par-4
Natural proteins in general fall into one of three categories: properly folded and functional, misfolded and nonfunctional, or functional despite lacking a stable folded structure. Intrinsically disordered proteins (IDPs), such as Par-4, fall into the latter group, as they do not adopt a fixed three-dimensional conformation and are considered natively unfolded. Rather than forming stable secondary, tertiary, or quaternary structures, IDPs exist as dynamic, flexible chains that fluctuate between conformations [90]. Some proteins also contain intrinsically disordered protein regions (IDPRs) within otherwise structured domains. Both IDPs and IDPRs play essential roles in cellular signaling and are more prevalent in eukaryotic organisms.
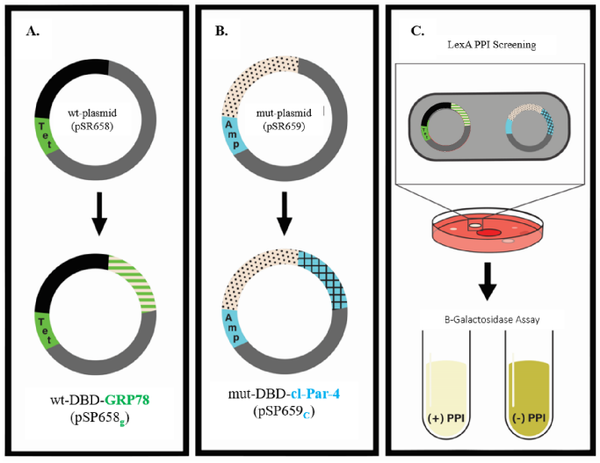
Schematic overview of a LexA-B2H assay
(A) GRP78 (green) was subcloned into the reporter vector carrying the wild-type LexA DBD to express wt-DBD-GRP78 fusion protein (FP); (B) cl-Par-4 (blue) was subcloned into a reporter vector carrying the mutant LexA DBD to express a mut-DBD-cl-Par-4 FP. (C) These two constructs were co-transformed into SU202 E. coli reporter cells, which were then plated onto MacConkey agar for pink and white colony screening. White colonies, indicating GRP78 and cl-Par-4 interaction, were sub-cultured for downstream β-galactosidase assays to quantify binding strength. This colorimetric assay measures β-galactosidase activity, producing a yellow product in the absence of a PPI, while a decrease in color indicates the presence of a PPI.

SDS-PAGE and Western blot analysis
(A) Proteins were separated by SDS-PAGE and stained with Coomassie blue to visualize total protein expression. FPs containing GRP78 (90 kDa) and cl-Par-4 (40 kDa) are observed at the expected sizes. (B) Western blot analysis. An anti-LexA antibody confirmed expression of FPs containing GRP78 (90 kDa) and cl-Par-4 (40 kDa) in lane 2. Lane 1 is a size ladder.
Assessment of PPIs using MacConkey Agar
(A-F) Negative controls produced dark pink colonies, confirming the absence of PPIs. (G-H) Positive controls formed pale white-pink colonies, indicating strong PPIs. (I) SU202 cells co-expressing wt-DBD-GRP78 FP and mut-DBD-cl-Par-4 FP generated pale pink colonies, suggesting a
strong interaction.

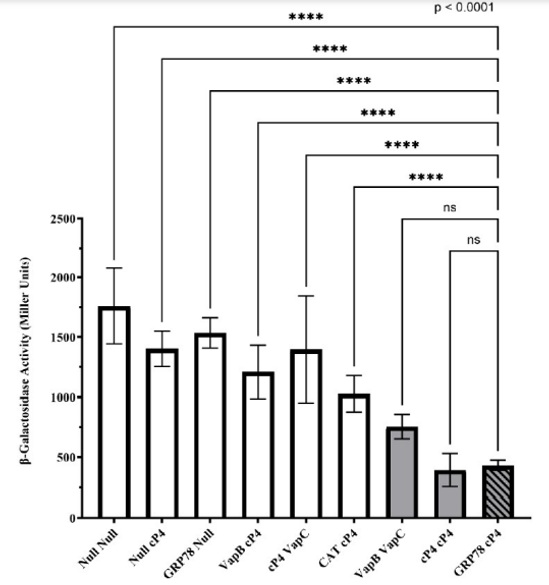
Quantitative Analysis of PPIs Using β-Galactosidase Assay
Negative controls (white bars) displayed high enzyme activity, ranging from 1029 to 1760 Miller units. In contrast, the two positive controls (gray bars) showed reduced activity, with values of 756 ± 102 and 394 ± 139 Miller units. The GRP78 and cl-Par-4 interaction resulted in 434 ± 41 Miller units, suggesting a binding affinity comparable to cl-Par-4 dimerization. All control groups were compared to the GRP78/cl-Par-4 test sample. An ordinary one-way ANOVA followed by Dunnett’s multiple comparisons test was conducted using GraphPad Prism (version 10.4.2). A significance threshold of p < 0.05 was used, and data are reported as mean ± standard deviation. Each dataset represents seven independent trials performed in triplicate.